
PDF) Comparison between parallel and distributed molecular dynamics simulations of Lennard-Jones systems | Dorian Gorgan - Academia.edu

A GPU-Accelerated Machine Learning Framework for Molecular Simulation: HOOMD-blue with TensorFlow | Theoretical and Computational Chemistry | ChemRxiv | Cambridge Open Engage

Fast and Flexible GPU Accelerated Binding Free Energy Calculations within the AMBER Molecular Dynamics Package | bioRxiv

Lennard-Jones Potentials for the Interaction of CO2 with Five-Membered Aromatic Heterocycles | The Journal of Physical Chemistry A

PDF) A GPU Accelerated Lennard-Jones System for Immersive Molecular Dynamics Simulations in Virtual Reality
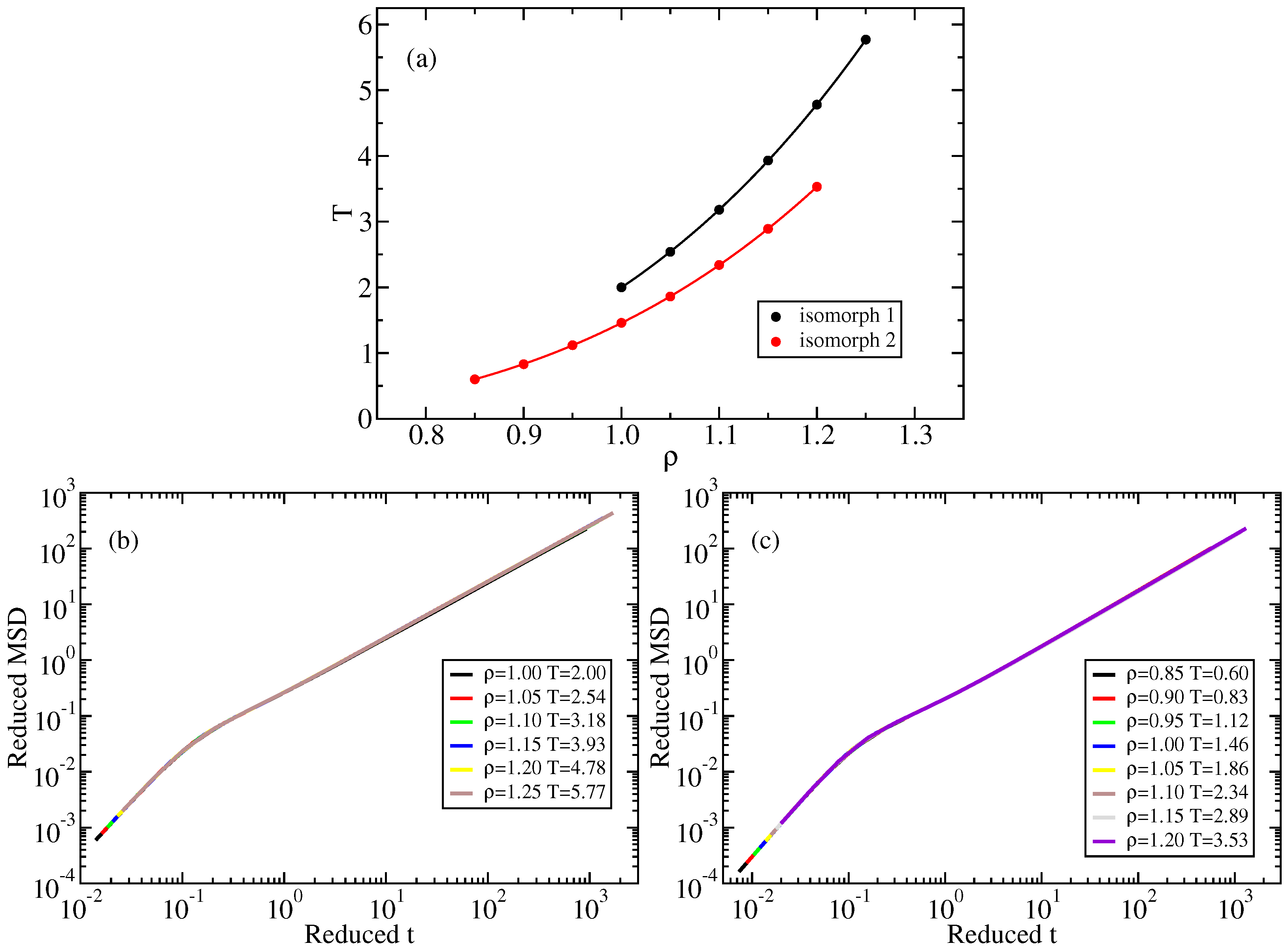

![exercises:2017_ethz_mmm:pythonmd [CP2K Open Source Molecular Dynamics ] exercises:2017_ethz_mmm:pythonmd [CP2K Open Source Molecular Dynamics ]](https://www.cp2k.org/_media/exercises:2017_ethz_mmm:screen_shot_2017-03-03_at_05.47.25.png?w=600&tok=47f5d6)

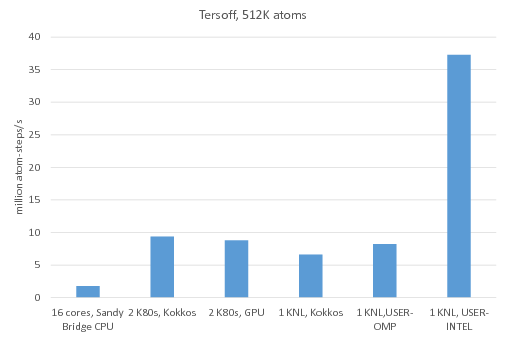




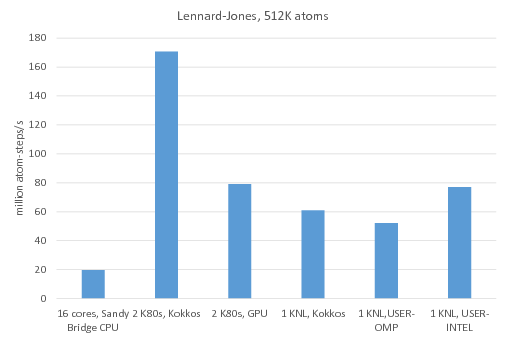
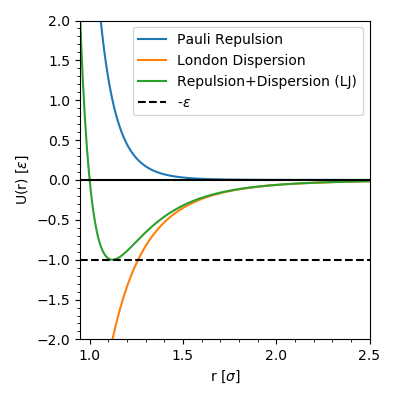
![PDF] GPU-accelerated Gibbs ensemble Monte Carlo simulations of Lennard-Jonesium | Semantic Scholar PDF] GPU-accelerated Gibbs ensemble Monte Carlo simulations of Lennard-Jonesium | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/76d4df3af9b558d2206a300e6803e9afabe28bb9/5-Figure4-1.png)
![PDF] Strong scaling of general-purpose molecular dynamics simulations on GPUs | Semantic Scholar PDF] Strong scaling of general-purpose molecular dynamics simulations on GPUs | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/f1d346ee291314ee2ab80c1d6930f5341a97264c/26-Figure14-1.png)
